Active
View Entry | In vitro assembly of a cell-signaling pathway3787T cells are white blood cells that are important in defending the body against bacteria, viruses and other pathogens. Each T cell carries proteins, called T-cell receptors, on its surface that are activated when they come in contact with an invader. This activation sets in motion a cascade of biochemical changes inside the T cell to mount a defense against the invasion.
Scientists have been interested for some time what happens after a T-cell receptor is activated. One obstacle has been to study how this signaling cascade, or pathway, proceeds inside T cells.
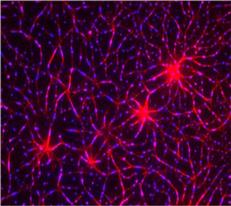
In this image, researchers have created a T-cell receptor pathway consisting of 12 proteins outside the cell on an artificial membrane. The image shows two key steps during the signaling process: clustering of a protein called linker for activation of T cells (LAT) (blue) and polymerization of the cytoskeleton protein actin (red). The findings show that the T-cell receptor signaling proteins self-organize into separate physical and biochemical compartments. This new system of studying molecular pathways outside the cells will enable scientists to better understand how the immune system combats microbes or other agents that cause infection.
To learn more how researchers assembled this T-cell receptor pathway, see this press release from HHMI's Marine Biological Laboratory Whitman Center. Related to video 3786. | | Public Note | | | | | Internal Note | | Researchers supplied image and gave permission for public use:
From: Michael Rosen [Michael.Rosen@UTSouthwestern.edu]
To: Spiering, Martin (NIH/NIGMS) [C]
Cc: Ron Vale ?[Ron.Vale@ucsf.edu]??; Su, Xiaolei ?[Xiaolei.Su@ucsf.edu]??; Jonathon Ditlev ?[Jonathon.Ditlev@UTSouthwestern.edu]?
Dear Martin,
Sorry for my mistake on my R01. The grant was just renewed, and I changed its focus to the actin dynamics work. The previous cycle did support some of the work in the Science paper. So you should feel free to use it.
Yours,
Mike
From: Michael Rosen [Michael.Rosen@UTSouthwestern.edu]
To:Vale, Ron ?[Ron.Vale@ucsf.edu]?
Cc: Spiering, Martin (NIH/NIGMS) [C]?; Su, Xiaolei ?[Xiaolei.Su@ucsf.edu]??; Jonathon Ditlev ?[Jonathon.Ditlev@UTSouthwestern.edu]?
Dear Martin,
Thanks for your interest in our work, which I'd be very happy to have in the NIGMS image gallery. My portion of the work was funded by HHMI, a grant from the Welch Foundation (#I-1544), and an F32 postdoctoral fellowship to Jon Ditlev from NIGMS. I'm cc:ing Jon here, and he can give you the grant number on the F32. Like Ron, my NIH R01 grant funds different work in my lab, and is currently focused on understanding the dynamics of actin using NMR spectroscopy.
Thanks again,
Mike | | | Keywords | | immune system, immune response | | | Source | | Xiaolei Su, HHMI Whitman Center of the Marine Biological Laboratory | | | Date | | | | | Credit Line | | Xiaolei Su, HHMI Whitman Center of the Marine Biological Laboratory | | | Investigator | | Michael Rosen, HHMI University of Texas | | | Record Type | | Photograph | | | Topic Area(s) | | ;#Cells;#Chemistry, Biochemistry, and Pharmacology;# | | | Previous Uses | | | | | Status | | Active | |
| | View All Properties | | Edit Properties |
|